U54 Center for Cancer Systems Biology
Despite anti-hormone therapies in patients, the cognate receptors ERα and AR can remain functional to support oncogenic signaling for advanced progression of breast and prostate cancers. Intensive studies have uncovered cellular and biochemical changes underlying the development of hormone resistance. However, epigenetic mechanisms for establishing and maintaining a hormone-resistant phenotype remain to be explored. Our recent studies have found remarkably similar epigenetic machinery that regulates hormone-independent gene transcription in both breast and prostate cancers. This process has multifaceted components, involving trans- and cis-acting elements, nucleosome reorganization, and chromatin interactions. To understand this complex mechanism, the San Antonio-Duke University Research Center for Cancer Systems Biology (SA-Duke RCCSB) has assembled a team of 21 experimental and computational investigators, and oncologists who plan to study a three-tiered epigenetic framework for gene regulation.
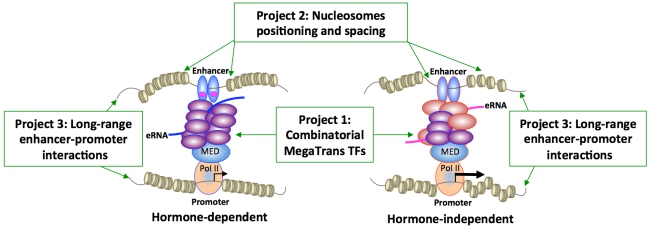
First, microenvironmental cues initiate the recruitment of a specific combination of trans-bound transcription factors (TFs), called MegaTrans TFs, to ERα or AR-bound enhancers (Project 1). MegaTrans TFs are composed of diverse signaling-dependent transcription factors that activate these enhancers through receiving other signal cues without hormone stimulation. Second, this hormone-independent action requires well-orchestrated repositioning of nucleosomes, enabling maximal MegaTrans-DNA contact in target chromatin regions (Project 2). Pioneer factor FOXA1 and chromatin remodelers are also critical regulators of repositioned nucleosomes during the transition of a hormone-sensitive to -resistant phenotype. Third, this concerted action triggers chromatin movement, remotely bringing the MegaTrans/enhancer complexes in close proximity to target promoters (Project 3). Intra- and inter-chromatin interactions facilitate the formation of transcriptional architectures that efficiently and autonomously regulate ERα/AR-mediated gene expression even in the absence of agonists or in the presence of antagonists. Experimental investigators plan to use omics-seq platforms to map combinatorial MegaTrans complexes, repositioned nucleosomes, and topologically associated domains (TADs) that spatiotemporally regulate hormone-independent transcription. Computational scientists then use omics data to derive 3D models of DNA-eRNA-protein interacting units in subnuclear compartments of cancer cells. Back to the bench, experimental scientists will use in silico findings to validate enhancer/gene markers that predict a hormone-resistant phenotype in patient-derived xenografts (PDXs) and clinical samples. To ensure seamless data integration of the three projects, a Data Analysis and Management Core will implement customized toolkits to manage computational infrastructure and store omics-seq metadata for heuristic queries by community systems biologists. An Outreach Core will facilitate the training of new-generation systems biologists and enhance collaborative efforts within the NCI’s consortium and in the 4D nucleosome community. An Administrative Core will provide governance and oversee rigorous evaluations of Intra-center Pilot Projects (IPPs), ensure cross-pollination between the bench and in silico scientists in the SA-Duke RCCSB, and reinforce national guidelines of data sharing.

